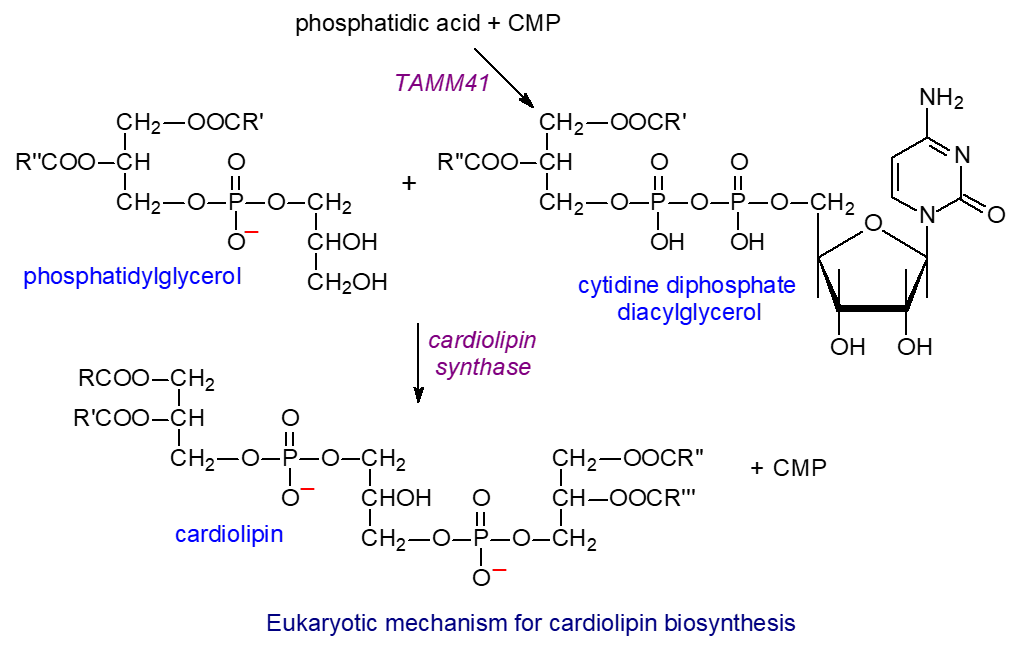
2. Virion and ribonucleoprotein
Coronaviruses are members of family Coronaviridae, order Nidovirales. These enveloped viruses possess genomes in the form of single-stranded RNA molecules of positive sense, that is, the same sense as the messenger RNA (mRNA). At present, four genera are known: Alphacoronavirus, Betacoronavirus, Gammacoronavirus, Deltacoronavirus. Members of the genera Alphacoronavirus and Betacoronavirus are identified to cause human disease, whereas those of the genera Gammacoronavirus and Deltacoronavirus are causative agents of animal disease .
Coronaviruses have a typical characteristic in negative-stained electron microscopy showing a fringe on their surface structure like a spike. This fringe resembles the solar corona, from which the name coronavirus was derived . These viruses are roughly spherical with average diameter of 80–120 nm. The surface spikes of the coronaviruses projects about 17–20 nm from the surface of the virus particle and have been described as club-like, pear-shaped, or petal-shaped, having a thin base which swells to a width of approximately 10 nm at the distal extremity . A schematic visualization of the coronavirus virion is presented in . In infection, the coronavirus particle serves three important functions for the genome: first, it provides the means to deliver the viral genome across the plasma membrane of a host cell; second, it serves as a means of escape for the newly synthesized genome; third, the viral particle functions as a durable vessel which protects the genome integrity on its journey between cells.

Figure 1.
Schematic diagram of the coronavirus virion. Together with the membrane (M) and envelope (E) transmembrane proteins, the spike (S) glycoprotein projects from a host cell-derived lipid bilayer, giving the virion a distinctive appearance. The haemagglutinin esterase (HE) forms small spikes which appear under the tall S protein spikes. The positive-sense viral genomic RNA is associated with the nucleocapsid phosphoprotein (N) forming the ribonucleoprotein with a helical structure
The genome of the coronaviruses codes four main structural proteins: the spike (S) protein, the nucleocapsid (N) protein, the membrane (M) protein and the envelope (E) protein, each of which play primary roles in the structure of the virus particle as well as in other aspects of the viral replication cycle. Generally, all of these proteins are needed to form a structurally complete virion. Some coronaviruses, however, do not require the full assemblage of the structural proteins to produce a complete, infectious viral particle. This indicates that some structural proteins are likely dispensable, or that those viruses may encode additional proteins with compensatory roles. The envelope of coronaviruses contains three or four viral proteins. The major proteins of the viral envelope are the S and the M proteins. In some, but not all coronaviruses, a third major envelope protein, the hemagglutinin esterase (HE) is found. Lastly, the small E protein constitutes a minor however critical structural component of the viral envelope. Many of the coronavirus proteins are modified by post-translational modifications which change the protein structure by proteolytic cleavage and disulfide bond formation or extend the chemical repertoire of the 20 standard amino acids by introducing new functional groups. Functional groups are commonly added through phosphorylation, glycosylation and lipidation (such as palmitoylation and myristoylation). The post-translational modifications play critical roles in regulating folding, stability, enzymatic activity, subcellular localization and interaction of the viral protein with other proteins.
In contrast to the other main structural proteins, the N protein is the only protein which mainly plays roles to bind to the viral RNA genome to form the nucleoprotein. However, apart from its primarily function in packaging and stabilizing the viral genome, the N protein also plays roles in other aspects of the coronavirus replication cycle and in the modulation of host cellular response to viral infection such as regulating the host cell cycle, affecting cell stress response, influencing the immune system, etc. Although the N protein is not required for the viral envelope formation, it may be required for the whole virion formation as transient expression of the gene encoding the N protein significantly increases the production of virus-like particles in some coronaviruses. The coronavirus has a large-sized genome, while the overall size of the viral particle is similar to that of other RNA viruses. It seems therefore that the space inside the coronavirus envelope would not be adequate to encapsulate loosely packed ribonucleoproteins. Surprisingly, the way the coronaviruses package their large genome is similar to that of the eukaryotic cells, that is in the form of a supercoiled dense structure. The incorporation of the coronavirus genomic RNA into a virion is dependent on the N proteins. Recent studies using mouse hepatitis virus (MHV)-infected cells showed that the cytoplasmic N proteins constitutively form oligomers through a process that does not need binding to genomic RNA. It was hypothesized that constitutive N protein oligomerization allows the optimal loading of the genomic viral RNA into a ribonucleoprotein complex through the presentation of multiple viral RNA binding motifs.
3. Spike (S) protein
The coronavirus spike (S) protein is a large glycosylated transmembrane protein ranging from about 1162 to 1452 amino acid residues. Monomers of the S protein, prior to glycosylation, are 128–160 kDa, but molecular masses of the glycosylated forms of the full-length monomer are 150–200 kDa. Following translation, the proteins fold into a metastable prefusion form and assemble into a homotrimer forming the coronavirus distinctive surface spike of crown-like appearance. The S protein is the most outward envelope protein of the coronaviruses. The S glycoprotein plays critical roles in mediating virus attachment to the host cell receptors and facilitating fusion between viral and host cell membranes.
figure 2
The S2 subunit of coronaviruses is highly conserved and contains segments that have critical roles to facilitate virus-cell fusion. These segments include the fusion peptide (FP), two heptad repeat regions, the heptad repeat region 1 (HR1 or HR-N), heptad repeat region 2 (HR2 or HR-C) and the highly conserved transmembrane domain.
4. Membrane (M) protein
The membrane (M) glycoprotein is the most abundant envelope protein of coronaviruses playing critical roles in the virion assembly through M-M, M-spike (S), and M-nucleocapsid (N) protein interactions. Generally, its length is 217–230 amino acids. It is a triple-spanning membrane protein with a short amino-terminal domain located on the exodomain of the virus (in the virion exterior, equivalent to the lumen of intracellular organelles) and a long carboxy-terminal domain in the endodomain of the virion (in the virion interior, equivalent to the cytoplasmic space of intracellular membranes). The nascent polypeptides, in the glycosylated forms, are of 25–30 kDa (221–262 amino acids) and the detected glycosylated forms are of higher molecular weights. The C-terminal domains of the MERS-CoV and IBV M proteins have been shown to contain signals for the trans-Golgi network and the endoplasmic reticulum-Golgi intermediate compartment (ERGIC)/cis-Golgi localization, of host cells respectively.
The M proteins from different coronaviruses show the same overall basic structure although their amino acid contents vary. The proteins have three transmembrane (TM) domains flanked by the amino terminal glycosylated domain and the carboxy-terminal domain. Multiple M domains and residues have been indicated to be essential for coronavirus assembly. After the third TM domain, the long intravirion (cytoplasmic) tail of M protein harbors an amphipathic domain and a short hydrophilic region at the carboxyl end of the tail. The amphipathic domain is suggested to be closely associated with the membrane. At the amino terminus of the amphipathic domain, there is a highly conserved 12-amino-acid domain with amino acid sequence SMWSFNPETNIL in the SARS-CoV M protein. This conserved domain (CD) has been suggested to be functionally important for M protein to participate in virus assembly. The schematic domain and membrane topology of the M protein is shown in
.
The schematic domain and membrane topology of the coronavirus membrane (M) protein. a). The coronavirus M protein has three transmembrane (TM) domains flanked by the amino terminal domain and the carboxy-terminal domain. The carboxy-terminal endodomain contains a conserved domain (CD) following the third transmembrane (TM) domain. b). The transmembrane topology of the coronavirus M protein. The M protein spans the viral membrane three times. The three transmembrane (TM) domains are flanked by the amino-terminal glycosylated domain (in the virion exterior) and the carboxy-terminal endodomain (in the virion interior). The conserved domain (CD) in the long carboxy-terminal endodomain is indicated.
5. Envelope (E) protein
The envelope (E) protein is a small integral membrane polypeptide, ranging from 76 to 109 amino acid residues with molecular weight of 8.4–12 kDa. The E protein plays important roles in a number of aspects of the coronavirus replication cycle, such as assembly, budding, envelope formation, and pathogenesis. Interestingly, although the protein is highly expressed inside the infected cells, only a small portion of the protein is incorporated into the viral envelope. Consequently, the protein is only a small constituent of the virus particle. Due to its small size and limited quantity, the E protein was identified much later compared to the other coronavirus structural proteins. Its primary and secondary structure indicates that the E protein has a short hydrophobic N terminus of 7–12 amino acid residues, followed by a transmembrane domain (TMD) of 25 amino acids, and ends with a long hydrophilic carboxy terminus. The E protein harbors conserved cysteine residues in the hydrophilic region that are targets for palmitoylation. In addition, it contains conserved proline residues in the C-terminal tail ()

. The schematic domain and membrane topology of coronavirus envelope (E) protein. a). The schematic domain of the coronavirus E protein. The protein has a hydrophobic domain predicted to span the viral membrane. The conserved cysteine and proline residues are indicated. b). Membrane topology of coronavirus E protein. The protein spans the viral membrane once with the N terminal end at the virion exterior and the C terminal end at the virion interior. The transmembrane domain is indicated by bar
6. Nucleocapsid (N) protein
The coronavirus nucleocapsid (N) protein is a structural phosphoprotein of 43–46 kDa, a component of the helical nucleocapsid. The main function of the N protein is to package the viral genome into a ribonucleoprotein (RNP) particle in order to protect the genomic RNA and for its incorporation into a viable virion. The N protein is thought to bind the genomic RNA in a beads-on-a-string fashion. In addition, it also interacts with the viral membrane protein during virion assembly and plays a critical role in improving the efficiency of virus transcription and assembly. The N protein undergoes rapid phosphorylation following its synthesis. In mouse hepatitis virus (MHV), phosphorylation occurs exclusively on serine residues. In infectious bronchitis virus (IBV), however, phosphorylation also takes place on threonine residues. The role of phosphorylation is unclear but it has been hypothesized to have a regulatory significance. The 46 kDa N protein of the SARS-CoV shares 20%–30% identity with other coronavirus N proteins. It forms a dimer which constitutes the basic building block of the nucleocapsid through its C-terminus. The N protein is dynamically associated with the replication-transcription complexes
Based on amino acid sequence comparisons it has been shown that the coronavirus N proteins have three distinct and highly conserved domains, namely the N terminal domain (NTD), the linker region (LKR) and the C-terminal domain (CTD). The NTD is separated from the CTD by the LKR, also termed an intrinsically disordered middle region ().
. The schematic domain of coronavirus nucleocapsid (N) protein. The coronavirus N protein is a phosphoprotein of 422 amino acid residues (in SARS-CoV). The protein has three distinct and highly conserved domains, the N terminal domain (NTD), the linker region (LKR) and the C-terminal domain (CTD). The NTD is separated from the CTD by the LKR. All of the three domains have been shown to bind with viral RNA. The LKR contains a Ser/Arg-rich region (SR) which contains a number of putative phosphorylation sites. The nuclear localization signal (NLS) motifs are shown. The N-terminal arm (NA) and the C-terminal tail (CT) are shown.
7. Accessory proteins
All coronavirus genomes contain accessory genes interspersed among the canonical genes, replicase, S, E, M, N which vary from as few as one (HCoV-NL63) to as many as eight genes (SARS-CoV). These accessory proteins are dispensable for coronavirus replication, however, they may confer biological advantages for the coronaviruses in the environment of the infected host cells. Some accessory proteins have been shown to exhibit roles in virus-host interaction and seem to have functions in viral pathogenesis. For SARS-CoV, some of the accessory proteins have been shown to be able to influence the interferon signaling pathways and the generation of pro-inflammatory cytokines. The accessory proteins encoded by the coronaviruses that infect humans are listed in .
Table 2
Accessory proteins of human coronaviruses∗.
| Virus | Accessory genes (Proteins) |
|---|
| HCoV-229E | [rep]-[S]-4a,4b-[E]-[M]-[N] |
| HCoV-NL63 | [rep]-[S]-3-[E]-[M]-[N] |
| HCoV-HKU1 | [rep]-2(HE)-[S]-4-[E]-[M]-[N], 7b(I) |
| HCoV-OC43 | [rep]-2a-2b (HE)-[S]-5 (12.9k)-[E]-[M]-[N], 7b(I) |
| SARS-CoV | [rep]-[S]-3a,3b-[E]-[M]-6-7a,7b-8a,8b-[N], 9b(I) |
| MERS-CoV | rep]-[S]-3-4a,4b-5-[E]-[M]-8b-[N] |
| SARS-CoV-2 | [rep]-[S]-3a,3b [E]-[M]-6-7a,7b-8b-[N],9b,10 |
8. Genome
The genome of coronaviruses is a nonsegmented, single-stranded RNA molecule with positive sense (+ssRNA), which is, of the same sense as the mRNA. Structurally it is similar to most eukaryotic mRNAs, in having 5'caps and 3′ poly-adenine tails. One of the distinctive features of the coronavirus genome is its remarkably large size ranging from 26 to 32 kb. For comparison, this is approximately three times the size of alphavirus or flavivirus genomes and four times the size of picornavirus genomes. Indeed, the size of the coronavirus genomes is among the largest known viral genomic RNAs. The genomes contain multiple ORFs, encoding a fixed array of structural and nonstructural proteins, as well as a variety of accessory proteins which differ in number and sequence among the coronaviruses.
About two-thirds of the 5′-most end of the genome is occupied by two large overlapping open reading frames, ORF1a and ORF1b. There is a -1 frameshift between ORF1a and ORF1b, leading to the synthesis of two polypeptides, pp1a and pp1ab, which are further processed by the viral proteases into 16 nonstructural proteins (nsps) which form the coronavirus replicase-transcriptase complex. This complex is an assembly of viral and hosts cellular proteins, which facilitate the synthesis of the genome and subgenome-sized mRNAs in the infected cell. The replicase-transcriptase complex plays an important role to amplify the genomic RNA and synthesize subgenomic mRNAs. Amplification of the genomic RNA involves full-length negative-strand templates, while the synthesis of subgenomic mRNA involves subgenome length negative-strand templates. The 16 nsps consist of nsp1– nsp11 encoded in ORF1a and nsp12–16 encoded in ORF1b. Studies in MHV-A59 have suggested that these proteins have multiple enzymatic functions, including papain-like proteases (nsp3), adenosine diphosphate-ribose 1,9-phosphatase (nsp3), 3C-like cysteine proteinase (nsp5), RNA-dependent RNA polymerase (nsp12), superfamily 1 helicase (nsp13), exonuclease (nsp14), endoribonuclease (nsp15), and S-adenosylmethionine-dependent 29-O-methyl transferase (nsp16). The ORF1a and ORF1b have been targeted for molecular detection of coronaviruses.
The remaining about one-third of the genome clustered at the 3′ end is transcribed into a nested set of subgenomic RNAs which contain ORFs for the structural proteins: spike (S), envelope (E), membrane (M) and nucleoprotein (N) as well as a variable number of accessory proteins depending on the viruses. The genes of accessory proteins are interspersed among the structural protein genes. Interestingly, there is a conserved gene order in all members of the coronavirus family, 5′-replicase-S-E-M-N-3’. However, genetic engineering experiments suggested that this evolutionary native order is not essential for functionality . Additionally, the genome has a 5′ UTR (untranslated region), ranging from 210 to 530 nucleotides, and 3′ UTR, ranging from 270 to 500 nucleotides . The 5′ 350 nucleotides folds into a set of RNA secondary structures which are well conserved, and in the Betacoronaviruses, have been suggested to play a critical role in the discontinuous synthesis of subgenomic RNAs. These functionally important cis-acting elements extend the 3′ of the 5′UTR into ORF1a. All of the 3′UTRs have a 3′-terminal poly(A) tail. The 3′UTR is similarly conserved and harbors all of the cis-acting sequences necessary for viral replication. All of the mRNAs carry identical 70–90 nucleotide leader sequences at their 5′ ends . The organization of human-infecting coronavirus genomes is shown in
.
The schematic diagram of structure of the human-infecting coronavirus genomes. Each bar represents the genomic organization of each coronavirus. The genomic regions or open-reading frames (ORFs) are compared. The structural proteins, including spike (S), envelope (E), membrane (M) and nucleocapsid (N) proteins, as well as non-structural proteins translated from ORF 1a and ORF 1b and accessory proteins are indicated. The tags indicate the name of the ORFs. 5′UTR = 5′ untranslated region, 3′UTR = 3′ untranslated region, An = poly(A) tail.
9. The life cycle of coronaviruses
9.1. Viral entry and membrane fusion
The infection of coronaviruses is initiated by the binding of the virus particles to the cellular receptors which leads to viral entry followed by fusion of the viral and host cellular membranes (). The membrane fusion event allows the release of the viral genome into the host cells cytoplasm, a process known as uncoating, which makes the viral genome available for translation. Coronavirus entry is facilitated by the trimeric transmembrane spike (S) glycoprotein, which mediates receptor binding and fusion of the viral and host membranes. The interaction between the S protein and the cellular receptor is the main determinant of host species range and tissue tropism. The S1 subunit (domain) of the coronavirus S proteins plays an important role in mediating the S protein binding to the host receptor. This S1 subunit shows the most diversity among coronaviruses and partly accounts for the wide host range of this virus family. Coronaviruses show complex patterns regarding receptor recognition and the diversity of receptor usage is one of the most profound features of coronaviruses. The human cellular receptor for the coronaviruses is listed in .

The schematic diagram of coronavirus life cycle. The coronavirus infection is initiated by the binding of the virus particles to the cellular receptors leading to viral entry followed by the viral and host cellular membrane fusion. After the membrane fusion event, the viral RNA is uncoated in the host cells cytoplasm. The ORF1a and ORF1ab are translated to produce pp1a and pp1ab, which are subsequently processed by the proteases encoded by ORF1a to produce 16 non-structural proteins (nsps) which form the RNA replicase–transcriptase complex (RTC). This complex localizes to modified intracellular membranes which are derived from the rough endoplasmic reticulum (ER) in the perinuclear region, and it drives the generation of negative-sense RNAs ((–)RNAs) through both replication and transcription. During replication, the full-length (–)RNA copies of the genome are synthezied and used as templates for the production of full-length (+)RNA genomes. During transcription, a subset of 7–9 subgenomic RNAs, including those encoding all structural proteins, is produced through discontinuous transcription. In this process, subgenomic (–)RNAs are synthesized by combining varying lengths of the 3′end of the genome with the 5′ leader sequence necessary for translation. These subgenomic (–)RNAs are then transcribed into subgenomic (+)mRNAs. The subgenomic mRNAs are then translated. The generated structural proteins are assembled into the ribonucleocapsid and viral envelope at the ER–Golgi intermediate compartment (ERGIC), followed by release of the newly produced coronavirus particle from the infected cell
Table 3
Receptor of human pathogenic coronaviruses.
9.2. Replication of coronavirus genome
The replication of the coronavirus genome is viewed as the most fundamental aspect of the coronavirus biology. As the largest group of RNA virus, coronaviruses require an RNA synthesis machinery with the fidelity to faithfully replicate their RNA. Coronavirus replication is achieved by employing complex mechanisms involving various proteins encoded by both viral and host cell genomes. Evolutionary, the virus genome contains relatively constant replicative genes which are indispensable for viral replication. Despite undergoing high mutation rates, RNA viral genomes still encode proteins with arrays of conserved sequence motifs playing roles in facilitating their genome replication and expression. Such proteins include the RNA-dependent RNA polymerase (RdRp), RNA helicase, chymotrypsin-like proteases, papain-like proteases, and metal binding proteins. In coronavirus genomes, all of the genes encoding these proteins are located in the ORF1 strategically located at the 5′-most end of the genome. In addition, viruses also exploit cellular proteins for multiple purposes in their replication cycle, including the attachment and entry into the cells, the initiation and regulation of RNA replication and transcription, protein synthesis, and the assembly of progeny virions. For these purposes, viruses typically subvert the normal components of cellular RNA processing and translational machinery to play both integral and regulatory roles in the replication, transcription, and translation of the viral genomes.
Soon after the accomplishment of receptor binding and membrane fusion events which lead to the release and uncoating of the viral RNA genome, the genomic replication cycle is started. In line with all other positive (+)-stranded RNA viruses, a coronavirus replicates its genome through synthesis of a complementary negative (‒)-strand RNA using the genomic RNA as a template. Firstly, using a continuous transcription process, the genome-size positive (+) stranded RNA is used as a template to make the genome-size negative (‒)-stranded RNA which subsequently serves as a template for the synthesis of the genome-size positive (+) stranded RNA progenies. Astonishingly, a coronavirus also synthesizes a number of shorter negative (‒)-stranded RNA of various sizes through discontinuous transcription process. These subgenome-length negative (‒)-stranded RNA molecules subsequently serve as templates for producing a number of positive (+) stranded RNAs of various sizes, termed subgenomic RNAs. For examples, during replication of MHV-A59, six subgenomic mRNA molecules are produced. The coronavirus genome and subgenomic mRNAs share identical 3′ sequences and form a 3′ nested set of RNA molecules. Interestingly, only the ORF at the 5’ region of each subgenomic mRNA is translated into a unique protein. Notably, the positive strands (genomes and subgenomic mRNA) are produced in relatively large amounts compared to the negative strands of genome- and subgenome-length RNA which serve as templates for genome and subgenomic mRNA synthesis.
Similar to many other positive (+) sense RNA viruses, coronaviruses use proteolytic processing to control expression of their replicative protein machineries. The critical roles of the pp1a/pp1ab polyprotein processing in genomic replication of coronaviruses are demonstrated by the prevention of RNA biosynthesis by proteinase inhibitors blocking essential proteolytic cleavages. Based on their physiological role, coronavirus proteinases are classified into main proteinases and accessory proteinases. All coronaviruses encode one main proteinase.
9.3. Virion assembly and budding
One of the distinctive features of coronaviruses is the location of their virion assembly. For most enveloped viruses, virion assembly takes place at the host cells plasma membrane. For coronaviruses, however, virion budding and assembly occurs at the endoplasmic reticulum-Golgi intermediate compartment (ERGIC). Coronaviruses, therefore, obtain their membrane envelope from ERGIC.
In the presence of a great excess of subgenomic RNA species, coronaviruses have the ability to select the genomic positive (+) sense single stranded RNA to be packaged into assembled virions. This high degree of selectivity is mediated by the coronaviruses genomic PS, a critical element for genomic RNA packaging, originally identified in MHV. One of the most characterized PS elements, called psi, is located at the 5′ leader region of the HIV genome. Two viral proteins, the N protein and the M protein, have been suggested to play roles in recognizing the PS. The coronavirus N protein has two highly basic domains, the NTD and CTD, and a mostly acidic carboxy-terminal domain, termed N3 within the C-terminal tail (CT) (). The CTD and the N3 domains have been proposed to recognize the PS. In vivo studies of SARS-CoV have also indicated that both the N-terminal and C-terminal domains of the N protein are crucial for recognition in the packaging RNA.
References;
1. Artika IM, Dewantari AK, Wiyatno A. Molecular biology of coronaviruses: current knowledge. Heliyon. 2020 Aug;6(8):e04743. doi: 10.1016/j.heliyon.2020.e04743. Epub 2020 Aug 17. PMID: 32835122; PMCID: PMC7430346.